class: center, middle, inverse, title-slide # BCB420 - Computational Systems Biology ## Lecture 4 - Exploring the data and basics of Normalization ### Ruth Isserlin ### 2020-01-28 --- ## Before we start ## Assignment #1 * <font size=5> Due February 4th, 2020 @ 20:00 </font> ## What to hand in? * **html rendered RNotebook** - you should be able to submit this through quercus * Make sure the notebook and all associated code is checked into your github repo as I will be pulling all the repos at the deadline and using them to compile your code. - Your checked in code must replicate the handed in notebook. * **Do not check the data file into your repo!** - your code should download the data from GEO and generate a new, cleaned data file. * Document your work and your code directly in the notebook. * **Read the paper associated with your data!** * You are allowed to use helper functions or methods but make sure when you source those files the paths to them are relative and that they are checked into your repo as well. --- ## Lecture Overview ### Data Exploration * Data standards - what are they and what are they good for? ### Data Normalization * Why do we need to normalize out data? * What different types of distributions are there and why does this matter? ### Identifier Mapping * BiomaRt example --- ## Recap of Last class * We used the GEOmetadb Bioc package to query the GEO meta data to help find: * RNASeq data * human * dataset from within 5 years * related to ovarian cancer * supplementary file is counts * [GEOmetadb](https://www.bioconductor.org/packages/release/bioc/html/GEOmetadb.html) is a great package that basically indexes all the meta data in GEO which is very helpful when it isn't based on the strongest of controlled vocabularies. * I narrowed my search down to the GEO identifier - GSE70072 * So where do we start? -- <font size=8> Read the paper! </font> --- ### Standards * [Functional Genomics Data Society](http://fged.org/) * MIAME - **M**inimum **I**nformation **A**bout a **M**icroarray **E**xperiment * originally published in [Nature genetics in 2001](https://www.nature.com/articles/ng1201-365) and represented the guiding principles of the minimal information needed for submitting a dataset * MINSEQE - **M**inimum **I**nformation about a high-throughput **SEQ**uencing **E**xperiment -- * **GEO representation of MIAME and MINSEQE** <img src=./images/img_lecture4/MIAME_MINSEQE.png> --- ### Standards * [Functional Genomics Data Society](http://fged.org/) * MIAME - **M**inimum **I**nformation **A**bout a **M**icroarray **E**xperiment * originally published in [Nature genetics in 2001](https://www.nature.com/articles/ng1201-365) and represented the guiding principles of the minimal information needed for submitting a dataset * MINSEQE - **M**inimum **I**nformation about a high-throughput **SEQ**uencing **E**xperiment * **GEO representation of MIAME and MINSEQE** <img src=./images/img_lecture4/MIAME_MINSEQE_highlighted.png> --- ## NCBI GEO standards and services for microarray data * An interesting read! [Nat Biotechnol. 2006 Dec; 24(12): 1471–1472.](https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2270403/) <img src=https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2270403/bin/nihms41580f1.jpg> --- ### Highlights from NCBI GEO standards and services for microarray data the MIAME requirements have been criticized recently. The criticism stems, in part, from **different interpretations of the level of detail required to adequately report a microarray experiment**, and debates as to whether there is a genuine benefit to making microarray data public. "In June 2005, we released major database revisions that included specific provisions for all MIAME data elements. In 2006, mechanisms for provision of raw data were further streamlined, and several MIAME elements that were previously optional became mandatory. Yet, even with these advances, it is still possible for a submitter to supply data that do not strictly adhere to the MIAME requirements. The difficulty lies in the fact that MIAME is a **subjective set of guidelines** where the level of detail to report is open to interpretation and, thus, cannot be unequivocally validated or enforced by computational means." "each submission is inspected by curators for content integrity. GEO curators employ a pragmatic approach; we aim to ensure that sufficient information has been supplied to allow general interpretation of the experiment. Although encouraged, we have been less dogmatic with regards to provision of all-inclusive experimental protocols that would possibly permit practical replication of the entire experiment. Our reasoning is that provision of granulated experimental details adds a significant burden on the submitter, for (arguably) minimal real benefit for most end-users who are usually less concerned with this level of detail." ??? It more of a user driven resource. A little bit of the honour system - these are the guidelines and we hope that submitters will abide by them but no one is forcing them to follow them. If a reviewer has comments then submitter can easily make adjustments - but how often do you think reviewers go and download the data. --- ## Standards - Proteomics [Proteomics Standards Initiative](http://www.psidev.info/) - PSI <img src=./images/img_lecture4/psi.png> ??? point out: the number of different branches of PSI that there are. they have standards and guidelines as well as controlled vocabularies. As a previous submitter the process is not easy but as you will see later in the course when we look at protein protein interaction networks the psi-mi format is very reach and easy to use computational. --- ## PSI-MI controlled vocabulary example <img src=./images/img_lecture4/psi_cv.png> ??? Active community that is still adding and adapting their controlled vocab and formats. The base is mostly stable but as new techonologies arise or evolve they are incorporated. Why is the proteomics community so different from the RNA community? not sure. There is a different amount of work and coordination associated with the different frameworks. --- Dataset = GSE70072 -- Title of the paper - Apoptosis enhancing drugs overcome innate platinum resistance in CA125 negative tumor initiating populations of high grade serous ovarian cancer -- Very briefly in my own words: * This study investigates two cell populations that they isolated from high-grade serous ovarian cancer, CA125+ve cells and CA125-ve cells. * They hypothesize the CA125-ve cells are stem-like cells and are the cause for resurgence of cancer following treatment as the chemo drugs kills all the CA125+ve cells enriching for CA125-ve cells. * They further demonstrate how a combination of two chemo drugs given together could help to kill both CA125+ve and CA125-ve cells. <font size=8>What is the first thing that you might want to investigate or ask with the above summary?</font> --- ## CA125 From the paper: * CA125 (Muc16) is a cell surface glycoprotein * highly expressed in high grade serous ovarian cancer and found in blood of effected patients. * a biomarker of the disease From Uniprot: * [Muc16](https://www.uniprot.org/uniprot/Q8WXI7) is not effective for stage I of the diseases so it isn't a good general marker of the disease. * not specific enough for population screening * used for monitoring the disease. ??? Highlight the common problem of different names. CA125 is not the recognized symbol of the gene. Why are they using it? Probably historical. --- ## General experimental design <img src=https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4532886/bin/ncomms8956-f1.jpg width="80%"> <font size=1>Janzen DM, Tiourin E, Salehi JA, Paik DY, Lu J, Pellegrini M, Memarzadeh S. [An apoptosis-enhancing drug overcomes platinum resistance in a tumour-initiating subpopulation of ovarian cancer](https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4532886/). Nat Commun. 2015 Aug 3;6:7956. - Figure 1</font> --- ## Specific RNASeq experimental design <img src=./images/img_lecture4/RNASeq_design_fig3c.png> --- ### Specific RNASeq experimental design - Corrected 6 months after initial publication * Original publication - August 2015 <img src=./images/img_lecture4/RNASeq_design_fig3c.png width="75%"> * Corrigendum - February 2016 <img src=./images/img_lecture4/RNASeq_design_fig3c_redo.png width="75%"> --- ### Error! * 341,056 of the 730,620 expression values were inadvertently paired with incorrect gene names. * the DNA repair and apoptosis genes did **not** show a statistically significant expression difference as first reported. * Although, this error did not effect their conclusions on the different treatment models they introduced just the possible mechanism that they proposed. <font size=7>Lesson Learned - Identifer mapping is important and the basis for all down stream analysis so we need to get it right!</font> --- ## Data Exploration * First things first get the GEO description of your dataset. ```r gse <- getGEO("GSE70072",GSEMatrix=FALSE) kable(data.frame(head(Meta(gse))), format = "html") ``` <table> <thead> <tr> <th style="text-align:left;"> contact_address </th> <th style="text-align:left;"> contact_city </th> <th style="text-align:left;"> contact_country </th> <th style="text-align:left;"> contact_department </th> <th style="text-align:left;"> contact_email </th> <th style="text-align:left;"> contact_institute </th> </tr> </thead> <tbody> <tr> <td style="text-align:left;"> 610 Charles E Young Dr East </td> <td style="text-align:left;"> Los Angeles </td> <td style="text-align:left;"> USA </td> <td style="text-align:left;"> Obstetrics and Gynecology </td> <td style="text-align:left;"> smemarzadeh@mednet.ucla.edu </td> <td style="text-align:left;"> UCLA </td> </tr> </tbody> </table> ??? GSEMatrix parameter will only work for microarray data --- ## Information about Platform - GPL11154 ```r current_gpl <- names(GPLList(gse))[1] current_gpl_info <- Meta(getGEO(current_gpl)) ``` -- ```r current_gpl_info$title ``` ``` ## [1] "Illumina HiSeq 2000 (Homo sapiens)" ``` ```r current_gpl_info$last_update_date ``` ``` ## [1] "Mar 27 2019" ``` ```r current_gpl_info$organism ``` ``` ## [1] "Homo sapiens" ``` --- ## Information about Platform - GPL11154 ### Represent the same as above but using markdown * with embedded r (expression) surrounded by [grave accent](https://en.wikipedia.org/wiki/Grave_accent). Also see: [R markdown cheatsheet](https://rstudio.com/wp-content/uploads/2015/02/rmarkdown-cheatsheet.pdf) for good tips<br><br> **Platform title** : Illumina HiSeq 2000 (Homo sapiens)<br> **Submission data** : Nov 02 2010<br> **Last update data** : Mar 27 2019<br> **Organims** : Homo sapiens (taxid: 9606)<br> **Number of GEO datasets that use this techology** : 7774 <br> (code: length(current_gpl_info$series_id)) <br> **Number of GEO samples that use this technology** : 120075 <br> (code: length(current_gpl_info$sample_id)) <br> --- ### Get the expression Data ```r sfiles = getGEOSuppFiles('GSE70072') fnames = rownames(sfiles) # there is only one supplemental file ca125_exp = read.delim(fnames[1],header=TRUE, * check.names = FALSE) ``` -- Difference between: ```r read.table(fnames[1],header=TRUE,check.names=FALSE,nrows = 1)[1,1:4] ``` ``` ## ensembl75_id gname Pt.A.CA125- Pt.A.CA125+ ## 1 ENSG00000000003 TSPAN6 1858 1626 ``` And ```r read.table(fnames[1],header=TRUE,check.names=TRUE,nrows = 1)[1,1:4] ``` ``` ## ensembl75_id gname Pt.A.CA125. Pt.A.CA125..1 ## 1 ENSG00000000003 TSPAN6 1858 1626 ``` --- ```r kable(ca125_exp[1:15,1:5], format = "html") ``` <table> <thead> <tr> <th style="text-align:left;"> ensembl75_id </th> <th style="text-align:left;"> gname </th> <th style="text-align:right;"> Pt.A.CA125- </th> <th style="text-align:right;"> Pt.A.CA125+ </th> <th style="text-align:right;"> pt.B.CA125- </th> </tr> </thead> <tbody> <tr> <td style="text-align:left;"> ENSG00000000003 </td> <td style="text-align:left;"> TSPAN6 </td> <td style="text-align:right;"> 1858 </td> <td style="text-align:right;"> 1626 </td> <td style="text-align:right;"> 2711 </td> </tr> <tr> <td style="text-align:left;"> ENSG00000000005 </td> <td style="text-align:left;"> TNMD </td> <td style="text-align:right;"> 20 </td> <td style="text-align:right;"> 28 </td> <td style="text-align:right;"> 0 </td> </tr> <tr> <td style="text-align:left;"> ENSG00000000419 </td> <td style="text-align:left;"> DPM1 </td> <td style="text-align:right;"> 900 </td> <td style="text-align:right;"> 1090 </td> <td style="text-align:right;"> 746 </td> </tr> <tr> <td style="text-align:left;"> ENSG00000000457 </td> <td style="text-align:left;"> SCYL3 </td> <td style="text-align:right;"> 236 </td> <td style="text-align:right;"> 138 </td> <td style="text-align:right;"> 343 </td> </tr> <tr> <td style="text-align:left;"> ENSG00000000460 </td> <td style="text-align:left;"> C1orf112 </td> <td style="text-align:right;"> 216 </td> <td style="text-align:right;"> 184 </td> <td style="text-align:right;"> 326 </td> </tr> <tr> <td style="text-align:left;"> ENSG00000000938 </td> <td style="text-align:left;"> FGR </td> <td style="text-align:right;"> 0 </td> <td style="text-align:right;"> 13 </td> <td style="text-align:right;"> 7 </td> </tr> <tr> <td style="text-align:left;"> ENSG00000000971 </td> <td style="text-align:left;"> CFH </td> <td style="text-align:right;"> 97 </td> <td style="text-align:right;"> 975 </td> <td style="text-align:right;"> 157 </td> </tr> <tr> <td style="text-align:left;"> ENSG00000001036 </td> <td style="text-align:left;"> FUCA2 </td> <td style="text-align:right;"> 85 </td> <td style="text-align:right;"> 103 </td> <td style="text-align:right;"> 166 </td> </tr> <tr> <td style="text-align:left;"> ENSG00000001084 </td> <td style="text-align:left;"> GCLC </td> <td style="text-align:right;"> 550 </td> <td style="text-align:right;"> 567 </td> <td style="text-align:right;"> 247 </td> </tr> <tr> <td style="text-align:left;"> ENSG00000001167 </td> <td style="text-align:left;"> NFYA </td> <td style="text-align:right;"> 424 </td> <td style="text-align:right;"> 391 </td> <td style="text-align:right;"> 532 </td> </tr> <tr> <td style="text-align:left;"> ENSG00000001460 </td> <td style="text-align:left;"> STPG1 </td> <td style="text-align:right;"> 256 </td> <td style="text-align:right;"> 151 </td> <td style="text-align:right;"> 786 </td> </tr> <tr> <td style="text-align:left;"> ENSG00000001461 </td> <td style="text-align:left;"> NIPAL3 </td> <td style="text-align:right;"> 1043 </td> <td style="text-align:right;"> 816 </td> <td style="text-align:right;"> 1136 </td> </tr> <tr> <td style="text-align:left;"> ENSG00000001497 </td> <td style="text-align:left;"> LAS1L </td> <td style="text-align:right;"> 2837 </td> <td style="text-align:right;"> 2157 </td> <td style="text-align:right;"> 2183 </td> </tr> <tr> <td style="text-align:left;"> ENSG00000001561 </td> <td style="text-align:left;"> ENPP4 </td> <td style="text-align:right;"> 181 </td> <td style="text-align:right;"> 177 </td> <td style="text-align:right;"> 477 </td> </tr> <tr> <td style="text-align:left;"> ENSG00000001617 </td> <td style="text-align:left;"> SEMA3F </td> <td style="text-align:right;"> 37 </td> <td style="text-align:right;"> 69 </td> <td style="text-align:right;"> 58 </td> </tr> </tbody> </table> --- ## Cleaning the data * How many unique genes do we have? * Are there any non-genes in our dataset? If so what are they? * Can we exclude them? ### How many genes do we have measurments for? ```r dim(ca125_exp) ``` ``` ## [1] 63677 22 ``` -- Define the groups ```r #get the 2 and third token from the column names samples <- data.frame(lapply(colnames(ca125_exp)[3:22], FUN=function(x){unlist(strsplit(x, split = "\\."))[c(2,3)]})) colnames(samples) <- colnames(ca125_exp)[3:22] rownames(samples) <- c("patients","cell_type") samples <- data.frame(t(samples)) ``` --- ## Are any of genes duplicated? and Why? Get the summarized counts for each gene ```r summarized_gene_counts <- sort(table(ca125_exp$gname),decreasing = TRUE) ``` Only output those that are greater than 1 ```r kable(summarized_gene_counts[which(summarized_gene_counts>1)[1:10]], format="html") ``` <table> <thead> <tr> <th style="text-align:left;"> Var1 </th> <th style="text-align:right;"> Freq </th> </tr> </thead> <tbody> <tr> <td style="text-align:left;"> Y_RNA </td> <td style="text-align:right;"> 881 </td> </tr> <tr> <td style="text-align:left;"> snoU13 </td> <td style="text-align:right;"> 478 </td> </tr> <tr> <td style="text-align:left;"> U3 </td> <td style="text-align:right;"> 87 </td> </tr> <tr> <td style="text-align:left;"> U6 </td> <td style="text-align:right;"> 61 </td> </tr> <tr> <td style="text-align:left;"> SNORD112 </td> <td style="text-align:right;"> 50 </td> </tr> <tr> <td style="text-align:left;"> Metazoa_SRP </td> <td style="text-align:right;"> 44 </td> </tr> <tr> <td style="text-align:left;"> SNORA70 </td> <td style="text-align:right;"> 32 </td> </tr> <tr> <td style="text-align:left;"> U8 </td> <td style="text-align:right;"> 30 </td> </tr> <tr> <td style="text-align:left;"> SNORA31 </td> <td style="text-align:right;"> 27 </td> </tr> <tr> <td style="text-align:left;"> 5S_rRNA </td> <td style="text-align:right;"> 26 </td> </tr> </tbody> </table> --- ## What are those? * [Y_RNA](https://en.wikipedia.org/wiki/Y_RNA) - small non-coding RNAs * [SNOR](https://en.wikipedia.org/wiki/Small_nucleolar_RNA) - small nucleolar RNA * U - small nuclear RNA ## Do I need to filter them out? -- * At this point - **No** * Difference of opinion. * If we base our analysis on ensembl gene ids then they are unique elements and we don't have to worry about them. * Collapsing them might increase their overall counts * **But we do need to filter out genes that have low counts!** --- * According to the edgeR protocol - Filter weakly expressed and noninformative (e.g., non-aligned) features. * **In edgeR, it is recommended to remove features without at least 1 read per million in n of the samples, where n is the size of the smallest group of replicates.** * For our example dataset - there are 10 samples of each group so n=10 ```r #translate out counts into counts per million using the edgeR package function cpm cpms = cpm(ca125_exp[,3:22]) rownames(cpms) <- ca125_exp[,1] # get rid of low counts *keep = rowSums(cpms >1) >=3 ca125_exp_filtered = ca125_exp[keep,] ``` What does that do to the dataset? -- ```r dim(ca125_exp_filtered) ``` ``` ## [1] 19192 22 ``` ??? It is not important that the 10 samples that have at least 1 count per million are from the same condition, just that there are 10 of them. --- ### Does that solve some of duplicate issues? Get the summarized counts for each gene ```r summarized_gene_counts_filtered <- sort(table(ca125_exp_filtered$gname),decreasing = TRUE) ``` Only output those that are greater than 1 ```r kable(summarized_gene_counts_filtered[which(summarized_gene_counts_filtered>1)[1:10]], format="html") ``` <table> <thead> <tr> <th style="text-align:left;"> Var1 </th> <th style="text-align:right;"> Freq </th> </tr> </thead> <tbody> <tr> <td style="text-align:left;"> Y_RNA </td> <td style="text-align:right;"> 3 </td> </tr> <tr> <td style="text-align:left;"> CFB </td> <td style="text-align:right;"> 2 </td> </tr> <tr> <td style="text-align:left;"> CKS1B </td> <td style="text-align:right;"> 2 </td> </tr> <tr> <td style="text-align:left;"> CRYBG3 </td> <td style="text-align:right;"> 2 </td> </tr> <tr> <td style="text-align:left;"> GOLGA7B </td> <td style="text-align:right;"> 2 </td> </tr> <tr> <td style="text-align:left;"> LIMS3 </td> <td style="text-align:right;"> 2 </td> </tr> <tr> <td style="text-align:left;"> NPIPA7 </td> <td style="text-align:right;"> 2 </td> </tr> <tr> <td style="text-align:left;"> PDXDC2P </td> <td style="text-align:right;"> 2 </td> </tr> <tr> <td style="text-align:left;"> POLR2J4 </td> <td style="text-align:right;"> 2 </td> </tr> <tr> <td style="text-align:left;"> PSMA2 </td> <td style="text-align:right;"> 2 </td> </tr> </tbody> </table> --- ## Normalization * First steps in any analysis. * Why is it important? Why do we need to normalize our data? -- <br>How we think our experimental data should look - Bull's eye every time! <img src="images/img_lecture4/bullseye_1.png"> --- ## Normalization * First steps in any analysis. * Why is it important? Why do we need to normalize our data? <br>How the data actually looks: <img src="images/img_lecture4/bullseye_2.png"> --- ## Variation -- .pull-left[ **Technical Variation** ] .pull-right[ **Biological Variation** ] --- ## Variation .pull-left[ **Technical Variation** - caused by instrumental, experimental variation - samples run on different days by different people with slightly different reagents - Want to control for these factors as best we can but with larger experiments there will always be some level of variation - read depth, gene length, ... - This is what we want to correct for. ] .pull-right[ **Biological Variation** - came from a different sample or a different condition - These are the things we are **interested in** ] --- ## Data distributions * a sampling of data will form some sort of distribution, a mathematical means of representing it. * in our experimental methods, we are sampling a set of genes/proteins and associating some metric with that sampling. * There are many different types of well charecterized distributions, including: 1. Normal distribution 1. Bimodal distribution 1. Poisson distribution 1. Power log distribution --- ### Normal distribution Generate a set of 1000 randomly selected numbers from the normal distribution. ```r r <- rnorm(1000, mean=0, sd=1) ``` If we graph this random distribution it will look like: -- ```r hist(r,freq = FALSE,breaks = 30, xlim = c(-4, 4),ylim = c(0, 1), main = "Normal Distribution", xlab = "x",ylab = "f(x)", col = "yellow") ``` --- -- <!-- --> --- If we then grab 100 values equally spaced between -4 and 4 ```r hist(r,freq = FALSE,breaks = 30, xlim = c(-4, 4),ylim = c(0, 1), main = "Normal Distribution", xlab = "x",ylab = "f(x)", col = "yellow") x <- seq(-4, 4, length.out = 100) #add the density distribution points(x, dnorm(x), type = "l", lwd = 2, col="firebrick") ``` --- If we then grab 100 values equally spaced between -4 and 4 <!-- --> --- ### Distribution of our data - Boxplot ```r data2plot <- log2(cpm(ca125_exp_filtered[,3:22])) boxplot(data2plot, xlab = "Samples", ylab = "log2 CPM", las = 2, cex = 0.5, cex.lab = 0.5, cex.axis = 0.5, main = "CA125 RNASeq Samples") #draw the median on each box plot abline(h = median(apply(data2plot, 2, median)), col = "green", lwd = 0.6, lty = "dashed") ``` --- ### Distribution of our data - Boxplot <!-- --> --- ### Distribution of our data - Density plot ```r counts_density <- apply(log2(cpm(ca125_exp_filtered[,3:22])), 2, density) #calculate the limits across all the samples xlim <- 0; ylim <- 0 for (i in 1:length(counts_density)) { xlim <- range(c(xlim, counts_density[[i]]$x)); ylim <- range(c(ylim, counts_density[[i]]$y)) } cols <- rainbow(length(counts_density)) ltys <- rep(1, length(counts_density)) #plot the first density plot to initialize the plot plot(counts_density[[1]], xlim=xlim, ylim=ylim, type="n", ylab="Smoothing density of log2-CPM", main="", cex.lab = 0.85) #plot each line for (i in 1:length(counts_density)) lines(counts_density[[i]], col=cols[i], lty=ltys[i]) #create legend legend("topright", colnames(data2plot), col=cols, lty=ltys, cex=0.75, border ="blue", text.col = "green4", merge = TRUE, bg = "gray90") ``` --- ### Distribution of our data - Density plot 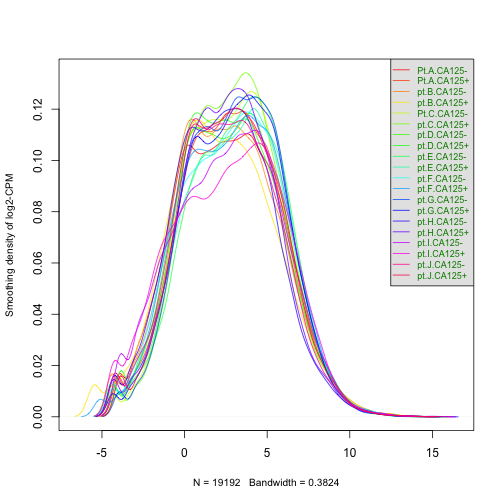<!-- --> --- ## Common Normalization methods ### Z-scored normalization * centres the data to zero and the standard deviation to 1. * assumes that the data is normally distributed ### Quantile normalization * sort each column by values * compute the average for each row * replace all values along the row with the computed average * put values back in original rows. * forces the distribution of each sample to be the same, the sum of each column is the same * **Most commonly used normalization method for microarray based technologies** <font size=6> What do these normalization methods lack with regards to RNASeq?</font> --- ## Specialized Normalization Techniques for RNASeq * both methods are very similar and give comparable results (according to in depth comparison that can be found [here](https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0206312)) ### Trimmed Mean of M-values (TMM) * implemented in the edgeR package * based on the hypothesis that most genes are not differentially expressed * Sample based ### Relative Log Expression (RLE) * implemented in the DESeq package * based on the hypothesis that most genes are not differentially expressed * gene based --- ## M vs A plot * As it is the premise of the TMM method, just briefly go over it. * Originally used in microarray experiments with two dye (Red vs green) to assay the technical variability between the intensities of the two dyes. * M - ratio, ratio Cy5(red) to Cy3(green), A - average of Cy5 and Cy3 * For RNASeq experiments it can be used to compare two samples or two groups of samples. ```r plotMA(log2(ca125_exp[,c(3,4)]), ylab="M - ratio log expression", main="CA125 + vs - - example") ``` <!-- --> --- ## Trimmed Mean of M-values * Trimmed mean - is the average after removing the upper and lower percentage of the data points. (By default, 30% of the M values and 5% of the A values) <img src=./images/img_lecture4/TMM_def.png> * N is the total reads for sample K, Y is the observed counts * TMM compares each sample to a reference. * Major difference to previous methods - the data does not need to be modified prior to normalization. --- ## Applying TMM to our dataset ### Create an edgeR container for RNASeq count data * use the filtered counts and make sure it is not a data.frame but is a matrix * give the defined groups that we are going to be comparing - cell type (CA125+ vs CA125-) ```r filtered_data_matrix <- as.matrix(ca125_exp_filtered[,3:22]) rownames(filtered_data_matrix) <- ca125_exp_filtered$ensembl75_id d = DGEList(counts=filtered_data_matrix, group=samples$cell_type) ``` ## Calculate the normalization factors ```r d = calcNormFactors(d) ``` --- * How did normalization change our data ```r #get the normalized data normalized_counts <- cpm(d) ``` <!-- --> --- * What happens when we use the same scale <!-- --> --- ### Distribution of our data - Boxplot <!-- --> --- Inspect the sample separation using a multidimenstional scaling plot or MDS plot post normalization. * A MDS plot represents the distances between samples ```r plotMDS(d, labels=rownames(samples), col = c("darkgreen","blue")[factor(samples$cell_type)]) ``` <!-- --> --- Next, estimate common and tagwise dispersion * dispersion is a parameter that describes how much your variance deviates from the mean. * this is specific to the edgeR pacakge and is used downstream when calculating differential expression. * we can estimate both the common and tagwise dispersion. Common dispersion calculates a common dispersion value for all genes, while the tagwise method calculates gene-specific dispersions. * Genes with more counts will have smaller variations between samples than genes with few counts. * dispersion squared is biological coeffient of variation (BCV) so dispersion is a measure of how much variation there is in your samples. using: ```r model_design <- model.matrix(~samples$patients + samples$cell_type+0) d <- estimateDisp(d, model_design) ``` --- Graphing the BCV * tagwise = genewise, each dot represents the BCV for a gene ```r plotBCV(d,col.tagwise = "black",col.common = "red") ``` <!-- --> --- Create a visual representation of the mean-variance relationship ```r *plotMeanVar(d, show.raw.vars = TRUE, show.tagwise.vars=FALSE, NBline=FALSE, show.ave.raw.vars = FALSE,show.binned.common.disp.vars = FALSE) ``` <!-- --> --- ```r plotMeanVar(d, show.raw.vars = TRUE, * show.tagwise.vars=TRUE, NBline=FALSE, show.ave.raw.vars = FALSE, show.binned.common.disp.vars = FALSE) ``` <!-- --> --- ```r plotMeanVar(d, show.raw.vars = TRUE, show.tagwise.vars=TRUE, * show.ave.raw.vars = TRUE, NBline=FALSE, show.binned.common.disp.vars = FALSE) ``` <!-- --> --- ```r plotMeanVar(d, show.raw.vars = TRUE, show.tagwise.vars=TRUE, show.ave.raw.vars = TRUE, NBline=FALSE, * show.binned.common.disp.vars = TRUE) ``` <!-- --> --- ```r plotMeanVar(d, show.raw.vars = TRUE, show.tagwise.vars=TRUE, show.ave.raw.vars = TRUE, * NBline=TRUE, show.binned.common.disp.vars = TRUE) ``` <!-- --> --- <img src="https://media.springernature.com/lw685/springer-static/image/art%3A10.1038%2Fnmeth.3963/MediaObjects/41592_2016_Article_BFnmeth3963_Fig1_HTML.jpg"> <font size=2>Wadi, L., Meyer, M., Weiser, J. et al. [Impact of outdated gene annotations on pathway enrichment analysis](https://www.nature.com/articles/nmeth.3963). Nat Methods 13, 705–706 (2016).</font> --- ## Identifier mapping **Why can't the data just contain the identifiers that are interested in, i.e genes?** #### Techonology * Often data generation works with different identifiers than what we need for the analysis. * Technology itself works on a different level and tracks different information so often aren't working gene level data. For example - * RNASeq deals with read counts of short fragments of RNA that don't necessarily map to an individual gene * Microarray deals with probes which are the base unit of analysis. At design phase probes are associated with different region of the genome but are not necessarily mapped to a gene. * Proteomics deals with peptides similar to the above examples. #### Changing annotations * Thankfully our knowledge of genes, gene regulation are constantly expanding. * New genes, regulatory mechnisms are being discovered * RNASeq data can be re analyzed given new information to uncover new things. (microarray not to same degree) --- ## Tools for Identifier Mapping * Depends on the data you are trying to map. * Often going to the source will result in the best mappping, for example, for best mapping of ensembl ids go to [Ensembl](https://useast.ensembl.org/index.html). Or if you want mapping for mouse genes go to Mouse Genome Informatics [MGI](http://www.informatics.jax.org/) * Popular Tools: * [Ensembl Biomart](https://www.ensembl.org/biomart/martview/5d0da2ec3ccb52f6596c568a5b574402) - has web interface as well as multiple packages - the bioc package [biomaRt](https://bioconductor.org/packages/release/bioc/html/biomaRt.html) and the R package [biomartr](https://academic.oup.com/bioinformatics/article/33/8/1216/2931816) which actually depends on the biomaRt bioc package * biomartr is an extension on the biomaRt package and is optimized for those trying to get annotation information out of biomart. * [BridgeDB](https://openrisknet.org/e-infrastructure/services/119/) - restful service with a bioc interface [tutorial](https://bioconductor.org/packages/devel/bioc/vignettes/BridgeDbR/inst/doc/tutorial.html) - flaky though * Wherever you get your mappings from it is important that you are working with up to date annotations and the right versions. --- ## Biomart * Let's check how many of my ensembl gene ids map to the same gene names associated with them. ## Get all the available marts ```r library(biomaRt) listMarts() ``` ``` ## biomart version ## 1 ENSEMBL_MART_ENSEMBL Ensembl Genes 99 ## 2 ENSEMBL_MART_MOUSE Mouse strains 99 ## 3 ENSEMBL_MART_SNP Ensembl Variation 99 ## 4 ENSEMBL_MART_FUNCGEN Ensembl Regulation 99 ``` --- ## What if you need a different or older verion. ```r listEnsemblArchives()[1:10,] ``` ``` ## name date url version ## 1 Ensembl GRCh37 Feb 2014 http://grch37.ensembl.org GRCh37 ## 2 Ensembl 99 Jan 2020 http://jan2020.archive.ensembl.org 99 ## 3 Ensembl 98 Sep 2019 http://sep2019.archive.ensembl.org 98 ## 4 Ensembl 97 Jul 2019 http://jul2019.archive.ensembl.org 97 ## 5 Ensembl 96 Apr 2019 http://apr2019.archive.ensembl.org 96 ## 6 Ensembl 95 Jan 2019 http://jan2019.archive.ensembl.org 95 ## 7 Ensembl 94 Oct 2018 http://oct2018.archive.ensembl.org 94 ## 8 Ensembl 93 Jul 2018 http://jul2018.archive.ensembl.org 93 ## 9 Ensembl 92 Apr 2018 http://apr2018.archive.ensembl.org 92 ## 10 Ensembl 91 Dec 2017 http://dec2017.archive.ensembl.org 91 ## current_release ## 1 ## 2 * ## 3 ## 4 ## 5 ## 6 ## 7 ## 8 ## 9 ## 10 ``` --- Connect to the desired mart ```r ensembl <- useMart("ensembl") ``` Get the set of datasets available ```r datasets <- listDatasets(ensembl) kable(head(datasets),format = "html") ``` <table> <thead> <tr> <th style="text-align:left;"> dataset </th> <th style="text-align:left;"> description </th> <th style="text-align:left;"> version </th> </tr> </thead> <tbody> <tr> <td style="text-align:left;"> acalliptera_gene_ensembl </td> <td style="text-align:left;"> Eastern happy genes (fAstCal1.2) </td> <td style="text-align:left;"> fAstCal1.2 </td> </tr> <tr> <td style="text-align:left;"> acarolinensis_gene_ensembl </td> <td style="text-align:left;"> Anole lizard genes (AnoCar2.0) </td> <td style="text-align:left;"> AnoCar2.0 </td> </tr> <tr> <td style="text-align:left;"> acchrysaetos_gene_ensembl </td> <td style="text-align:left;"> Golden eagle genes (bAquChr1.2) </td> <td style="text-align:left;"> bAquChr1.2 </td> </tr> <tr> <td style="text-align:left;"> acitrinellus_gene_ensembl </td> <td style="text-align:left;"> Midas cichlid genes (Midas_v5) </td> <td style="text-align:left;"> Midas_v5 </td> </tr> <tr> <td style="text-align:left;"> amelanoleuca_gene_ensembl </td> <td style="text-align:left;"> Panda genes (ailMel1) </td> <td style="text-align:left;"> ailMel1 </td> </tr> <tr> <td style="text-align:left;"> amexicanus_gene_ensembl </td> <td style="text-align:left;"> Mexican tetra genes (Astyanax_mexicanus-2.0) </td> <td style="text-align:left;"> Astyanax_mexicanus-2.0 </td> </tr> </tbody> </table> --- Limit to the human datasets available ```r kable(head(datasets[grep(datasets$dataset,pattern = "sapiens"),]),format = "html") ``` <table> <thead> <tr> <th style="text-align:left;"> </th> <th style="text-align:left;"> dataset </th> <th style="text-align:left;"> description </th> <th style="text-align:left;"> version </th> </tr> </thead> <tbody> <tr> <td style="text-align:left;"> 78 </td> <td style="text-align:left;"> hsapiens_gene_ensembl </td> <td style="text-align:left;"> Human genes (GRCh38.p13) </td> <td style="text-align:left;"> GRCh38.p13 </td> </tr> </tbody> </table> ```r ensembl = useDataset("hsapiens_gene_ensembl",mart=ensembl) ``` --- ## Building a Biomart Query Function that we will be using is: **getBM()** ```r help(getBM) ``` What are we trying to convert: * Human Ensembl Gene Ids to HGNC symbols * A few things we need: 1. attributes - the type of identifiers that you want to retrieve. **So in our use case this is Ensembl gene Ids and the HGNC symbols** - What would happen if we just specified the HGNC symbols here? 1. filters - how you are filtering your results. Not intuitive. **For our use case this is Ensembl gene ids** 1. values - the values associated with the filters. **So in our use case this is the set of Ensembl gene ids that we have** --- ### Filters * unfortunately we have to figure out the correct name for all the identifiers that we are using as well as converting to. how many filters are there? -- ```r dim(listFilters(ensembl)) ``` ``` ## [1] 434 2 ``` -- What do those filters look like? ```r kable(listFilters(ensembl)[1:10,1:2], type="html") ``` <table> <thead> <tr> <th style="text-align:left;"> name </th> <th style="text-align:left;"> description </th> </tr> </thead> <tbody> <tr> <td style="text-align:left;"> chromosome_name </td> <td style="text-align:left;"> Chromosome/scaffold name </td> </tr> <tr> <td style="text-align:left;"> start </td> <td style="text-align:left;"> Start </td> </tr> <tr> <td style="text-align:left;"> end </td> <td style="text-align:left;"> End </td> </tr> <tr> <td style="text-align:left;"> band_start </td> <td style="text-align:left;"> Band Start </td> </tr> <tr> <td style="text-align:left;"> band_end </td> <td style="text-align:left;"> Band End </td> </tr> <tr> <td style="text-align:left;"> marker_start </td> <td style="text-align:left;"> Marker Start </td> </tr> <tr> <td style="text-align:left;"> marker_end </td> <td style="text-align:left;"> Marker End </td> </tr> <tr> <td style="text-align:left;"> encode_region </td> <td style="text-align:left;"> Encode region </td> </tr> <tr> <td style="text-align:left;"> strand </td> <td style="text-align:left;"> Strand </td> </tr> <tr> <td style="text-align:left;"> chromosomal_region </td> <td style="text-align:left;"> e.g. 1:100:10000:-1, 1:100000:200000:1 </td> </tr> </tbody> </table> --- ### filters - cont'd ```r biomart_human_filters <- listFilters(ensembl) kable(biomart_human_filters[ grep(biomart_human_filters$name,pattern="ensembl"),], format="html") %>% row_spec(3, background = "yellow") ``` <table> <thead> <tr> <th style="text-align:left;"> </th> <th style="text-align:left;"> name </th> <th style="text-align:left;"> description </th> </tr> </thead> <tbody> <tr> <td style="text-align:left;"> 13 </td> <td style="text-align:left;"> with_clone_based_ensembl_gene </td> <td style="text-align:left;"> With Clone-based (Ensembl) gene ID(s) </td> </tr> <tr> <td style="text-align:left;"> 14 </td> <td style="text-align:left;"> with_clone_based_ensembl_transcript </td> <td style="text-align:left;"> With Clone-based (Ensembl) transcript ID(s) </td> </tr> <tr> <td style="text-align:left;background-color: yellow !important;"> 55 </td> <td style="text-align:left;background-color: yellow !important;"> ensembl_gene_id </td> <td style="text-align:left;background-color: yellow !important;"> Gene stable ID(s) [e.g. ENSG00000000003] </td> </tr> <tr> <td style="text-align:left;"> 56 </td> <td style="text-align:left;"> ensembl_gene_id_version </td> <td style="text-align:left;"> Gene stable ID(s) with version [e.g. ENSG00000000003.15] </td> </tr> <tr> <td style="text-align:left;"> 57 </td> <td style="text-align:left;"> ensembl_transcript_id </td> <td style="text-align:left;"> Transcript stable ID(s) [e.g. ENST00000000233] </td> </tr> <tr> <td style="text-align:left;"> 58 </td> <td style="text-align:left;"> ensembl_transcript_id_version </td> <td style="text-align:left;"> Transcript stable ID(s) with version [e.g. ENST00000000233.10] </td> </tr> <tr> <td style="text-align:left;"> 59 </td> <td style="text-align:left;"> ensembl_peptide_id </td> <td style="text-align:left;"> Protein stable ID(s) [e.g. ENSP00000000233] </td> </tr> <tr> <td style="text-align:left;"> 60 </td> <td style="text-align:left;"> ensembl_peptide_id_version </td> <td style="text-align:left;"> Protein stable ID(s) with version [e.g. ENSP00000000233.5] </td> </tr> <tr> <td style="text-align:left;"> 61 </td> <td style="text-align:left;"> ensembl_exon_id </td> <td style="text-align:left;"> Exon ID(s) [e.g. ENSE00000327880] </td> </tr> <tr> <td style="text-align:left;"> 67 </td> <td style="text-align:left;"> clone_based_ensembl_gene </td> <td style="text-align:left;"> Clone-based (Ensembl) gene ID(s) [e.g. ABBA01000933.1] </td> </tr> <tr> <td style="text-align:left;"> 68 </td> <td style="text-align:left;"> clone_based_ensembl_transcript </td> <td style="text-align:left;"> Clone-based (Ensembl) transcript ID(s) [e.g. ABBA01000933.1-201] </td> </tr> </tbody> </table> --- ### Attributes ```r kable(listAttributes(ensembl)[1:10,1:2], type="html") ``` <table> <thead> <tr> <th style="text-align:left;"> name </th> <th style="text-align:left;"> description </th> </tr> </thead> <tbody> <tr> <td style="text-align:left;"> ensembl_gene_id </td> <td style="text-align:left;"> Gene stable ID </td> </tr> <tr> <td style="text-align:left;"> ensembl_gene_id_version </td> <td style="text-align:left;"> Gene stable ID version </td> </tr> <tr> <td style="text-align:left;"> ensembl_transcript_id </td> <td style="text-align:left;"> Transcript stable ID </td> </tr> <tr> <td style="text-align:left;"> ensembl_transcript_id_version </td> <td style="text-align:left;"> Transcript stable ID version </td> </tr> <tr> <td style="text-align:left;"> ensembl_peptide_id </td> <td style="text-align:left;"> Protein stable ID </td> </tr> <tr> <td style="text-align:left;"> ensembl_peptide_id_version </td> <td style="text-align:left;"> Protein stable ID version </td> </tr> <tr> <td style="text-align:left;"> ensembl_exon_id </td> <td style="text-align:left;"> Exon stable ID </td> </tr> <tr> <td style="text-align:left;"> description </td> <td style="text-align:left;"> Gene description </td> </tr> <tr> <td style="text-align:left;"> chromosome_name </td> <td style="text-align:left;"> Chromosome/scaffold name </td> </tr> <tr> <td style="text-align:left;"> start_position </td> <td style="text-align:left;"> Gene start (bp) </td> </tr> </tbody> </table> --- ### Attributes * you can also use the function searchAttribtutes to find the attribute you are looking for. ```r kable(searchAttributes(mart = ensembl, 'hgnc') , format="html") %>% row_spec(2, background = "yellow") ``` <table> <thead> <tr> <th style="text-align:left;"> </th> <th style="text-align:left;"> name </th> <th style="text-align:left;"> description </th> <th style="text-align:left;"> page </th> </tr> </thead> <tbody> <tr> <td style="text-align:left;"> 63 </td> <td style="text-align:left;"> hgnc_id </td> <td style="text-align:left;"> HGNC ID </td> <td style="text-align:left;"> feature_page </td> </tr> <tr> <td style="text-align:left;background-color: yellow !important;"> 64 </td> <td style="text-align:left;background-color: yellow !important;"> hgnc_symbol </td> <td style="text-align:left;background-color: yellow !important;"> HGNC symbol </td> <td style="text-align:left;background-color: yellow !important;"> feature_page </td> </tr> <tr> <td style="text-align:left;"> 93 </td> <td style="text-align:left;"> hgnc_trans_name </td> <td style="text-align:left;"> Transcript name ID </td> <td style="text-align:left;"> feature_page </td> </tr> </tbody> </table> --- * if you wanted to look for all of them at once ```r kable(searchAttributes(mart = ensembl, 'ensembl|hgnc')[1:12,] , format="html") %>% row_spec(c(1,11), background = "yellow") ``` <table> <thead> <tr> <th style="text-align:left;"> </th> <th style="text-align:left;"> name </th> <th style="text-align:left;"> description </th> <th style="text-align:left;"> page </th> </tr> </thead> <tbody> <tr> <td style="text-align:left;background-color: yellow !important;"> 1 </td> <td style="text-align:left;background-color: yellow !important;"> ensembl_gene_id </td> <td style="text-align:left;background-color: yellow !important;"> Gene stable ID </td> <td style="text-align:left;background-color: yellow !important;"> feature_page </td> </tr> <tr> <td style="text-align:left;"> 2 </td> <td style="text-align:left;"> ensembl_gene_id_version </td> <td style="text-align:left;"> Gene stable ID version </td> <td style="text-align:left;"> feature_page </td> </tr> <tr> <td style="text-align:left;"> 3 </td> <td style="text-align:left;"> ensembl_transcript_id </td> <td style="text-align:left;"> Transcript stable ID </td> <td style="text-align:left;"> feature_page </td> </tr> <tr> <td style="text-align:left;"> 4 </td> <td style="text-align:left;"> ensembl_transcript_id_version </td> <td style="text-align:left;"> Transcript stable ID version </td> <td style="text-align:left;"> feature_page </td> </tr> <tr> <td style="text-align:left;"> 5 </td> <td style="text-align:left;"> ensembl_peptide_id </td> <td style="text-align:left;"> Protein stable ID </td> <td style="text-align:left;"> feature_page </td> </tr> <tr> <td style="text-align:left;"> 6 </td> <td style="text-align:left;"> ensembl_peptide_id_version </td> <td style="text-align:left;"> Protein stable ID version </td> <td style="text-align:left;"> feature_page </td> </tr> <tr> <td style="text-align:left;"> 7 </td> <td style="text-align:left;"> ensembl_exon_id </td> <td style="text-align:left;"> Exon stable ID </td> <td style="text-align:left;"> feature_page </td> </tr> <tr> <td style="text-align:left;"> 51 </td> <td style="text-align:left;"> clone_based_ensembl_gene </td> <td style="text-align:left;"> Clone-based (Ensembl) gene ID </td> <td style="text-align:left;"> feature_page </td> </tr> <tr> <td style="text-align:left;"> 52 </td> <td style="text-align:left;"> clone_based_ensembl_transcript </td> <td style="text-align:left;"> Clone-based (Ensembl) transcript ID </td> <td style="text-align:left;"> feature_page </td> </tr> <tr> <td style="text-align:left;"> 63 </td> <td style="text-align:left;"> hgnc_id </td> <td style="text-align:left;"> HGNC ID </td> <td style="text-align:left;"> feature_page </td> </tr> <tr> <td style="text-align:left;background-color: yellow !important;"> 64 </td> <td style="text-align:left;background-color: yellow !important;"> hgnc_symbol </td> <td style="text-align:left;background-color: yellow !important;"> HGNC symbol </td> <td style="text-align:left;background-color: yellow !important;"> feature_page </td> </tr> <tr> <td style="text-align:left;"> 93 </td> <td style="text-align:left;"> hgnc_trans_name </td> <td style="text-align:left;"> Transcript name ID </td> <td style="text-align:left;"> feature_page </td> </tr> </tbody> </table> --- What are we trying to convert: * Human Ensembl Gene Ids to HGNC symbols * A few things we need: 1. attributes - the type of identifiers that you want to retrieve. * ensembl_gene_id and hgnc_symbol 1. filters - how you are filtering your results. Not intuitive. * ensembl_gene_id 1. values - the values associated with the filters. * ensembl75_id - first column in our expression matrix ```r #check to see if ca125_id_conversion file exists (computationally intensive) conversion_stash <- "ca125_id_conversion.rds" if(file.exists(conversion_stash)){ ca125_id_conversion <- readRDS(conversion_stash) } else { ca125_id_conversion <- getBM(attributes = c("ensembl_gene_id","hgnc_symbol"), filters = c("ensembl_gene_id"), values = ca125_exp_filtered$ensembl75_id, mart = ensembl) saveRDS(ca125_id_conversion, conversion_stash) } ``` --- How did we do? ```r nrow(normalized_counts) - nrow(ca125_id_conversion) ``` ``` ## [1] 552 ``` -- That is the difference between the number of mappings we have and the number of rows in our normalized set but that doesn't necessarily mean that we are missing only that many. --- Instead, lets try and merge our new identifiers. ```r normalized_counts_annot <- merge(ca125_id_conversion,normalized_counts,by.x = 1, by.y = 0, all.y=TRUE) kable(normalized_counts_annot[1:5,1:5],type = "html") ``` <table> <thead> <tr> <th style="text-align:left;"> ensembl_gene_id </th> <th style="text-align:left;"> hgnc_symbol </th> <th style="text-align:right;"> Pt.A.CA125- </th> <th style="text-align:right;"> Pt.A.CA125+ </th> <th style="text-align:right;"> pt.B.CA125- </th> </tr> </thead> <tbody> <tr> <td style="text-align:left;"> ENSG00000000003 </td> <td style="text-align:left;"> TSPAN6 </td> <td style="text-align:right;"> 122.26258 </td> <td style="text-align:right;"> 99.3479813 </td> <td style="text-align:right;"> 141.8717847 </td> </tr> <tr> <td style="text-align:left;"> ENSG00000000419 </td> <td style="text-align:left;"> DPM1 </td> <td style="text-align:right;"> 59.22299 </td> <td style="text-align:right;"> 66.5985852 </td> <td style="text-align:right;"> 39.0395984 </td> </tr> <tr> <td style="text-align:left;"> ENSG00000000457 </td> <td style="text-align:left;"> SCYL3 </td> <td style="text-align:right;"> 15.52958 </td> <td style="text-align:right;"> 8.4317475 </td> <td style="text-align:right;"> 17.9498422 </td> </tr> <tr> <td style="text-align:left;"> ENSG00000000460 </td> <td style="text-align:left;"> C1orf112 </td> <td style="text-align:right;"> 14.21352 </td> <td style="text-align:right;"> 11.2423300 </td> <td style="text-align:right;"> 17.0601999 </td> </tr> <tr> <td style="text-align:left;"> ENSG00000000938 </td> <td style="text-align:left;"> FGR </td> <td style="text-align:right;"> 0.00000 </td> <td style="text-align:right;"> 0.7942951 </td> <td style="text-align:right;"> 0.3663233 </td> </tr> </tbody> </table> --- How many identifiers are missing? ```r ensembl_id_missing_gene <- normalized_counts_annot$ensembl_gene_id[ which(is.na(normalized_counts_annot$hgnc_symbol))] length(ensembl_id_missing_gene) ``` ``` ## [1] 553 ``` ```r kable(normalized_counts_annot[which(is.na(normalized_counts_annot$hgnc_symbol))[1:5],1:5], type="html") ``` <table> <thead> <tr> <th style="text-align:left;"> </th> <th style="text-align:left;"> ensembl_gene_id </th> <th style="text-align:left;"> hgnc_symbol </th> <th style="text-align:right;"> Pt.A.CA125- </th> <th style="text-align:right;"> Pt.A.CA125+ </th> <th style="text-align:right;"> pt.B.CA125- </th> </tr> </thead> <tbody> <tr> <td style="text-align:left;"> 103 </td> <td style="text-align:left;"> ENSG00000005955 </td> <td style="text-align:left;"> NA </td> <td style="text-align:right;"> 45.8649180 </td> <td style="text-align:right;"> 56.7004469 </td> <td style="text-align:right;"> 40.7142193 </td> </tr> <tr> <td style="text-align:left;"> 111 </td> <td style="text-align:left;"> ENSG00000006074 </td> <td style="text-align:left;"> NA </td> <td style="text-align:right;"> 0.1316067 </td> <td style="text-align:right;"> 0.7331954 </td> <td style="text-align:right;"> 0.0000000 </td> </tr> <tr> <td style="text-align:left;"> 112 </td> <td style="text-align:left;"> ENSG00000006075 </td> <td style="text-align:left;"> NA </td> <td style="text-align:right;"> 1.8424931 </td> <td style="text-align:right;"> 8.0651498 </td> <td style="text-align:right;"> 3.4539055 </td> </tr> <tr> <td style="text-align:left;"> 113 </td> <td style="text-align:left;"> ENSG00000006114 </td> <td style="text-align:left;"> NA </td> <td style="text-align:right;"> 11.7787953 </td> <td style="text-align:right;"> 10.9979315 </td> <td style="text-align:right;"> 8.8964232 </td> </tr> <tr> <td style="text-align:left;"> 340 </td> <td style="text-align:left;"> ENSG00000017621 </td> <td style="text-align:left;"> NA </td> <td style="text-align:right;"> 0.3948200 </td> <td style="text-align:right;"> 2.8716821 </td> <td style="text-align:right;"> 0.7326466 </td> </tr> </tbody> </table> --- ## What do we do with these missing identifiers? {~3% of our dataset} ```r old_mapping <- merge(ca125_exp[,1:2],data.frame(ensembl_id_missing_gene),all.y = TRUE, by.x=1, by.y=1) ``` ```r kable(old_mapping[1:10,], type="html") ``` <table> <thead> <tr> <th style="text-align:left;"> ensembl75_id </th> <th style="text-align:left;"> gname </th> </tr> </thead> <tbody> <tr> <td style="text-align:left;"> ENSG00000005955 </td> <td style="text-align:left;"> GGNBP2 </td> </tr> <tr> <td style="text-align:left;"> ENSG00000006074 </td> <td style="text-align:left;"> CCL18 </td> </tr> <tr> <td style="text-align:left;"> ENSG00000006075 </td> <td style="text-align:left;"> CCL3 </td> </tr> <tr> <td style="text-align:left;"> ENSG00000006114 </td> <td style="text-align:left;"> SYNRG </td> </tr> <tr> <td style="text-align:left;"> ENSG00000017621 </td> <td style="text-align:left;"> MAGIX </td> </tr> <tr> <td style="text-align:left;"> ENSG00000034063 </td> <td style="text-align:left;"> UHRF1 </td> </tr> <tr> <td style="text-align:left;"> ENSG00000049319 </td> <td style="text-align:left;"> SRD5A2 </td> </tr> <tr> <td style="text-align:left;"> ENSG00000056661 </td> <td style="text-align:left;"> PCGF2 </td> </tr> <tr> <td style="text-align:left;"> ENSG00000068793 </td> <td style="text-align:left;"> CYFIP1 </td> </tr> <tr> <td style="text-align:left;"> ENSG00000069712 </td> <td style="text-align:left;"> KIAA1107 </td> </tr> </tbody> </table> --- * There are 553 RP1 gene names ```r kable(old_mapping[grep(old_mapping$gname,pattern = "[^RP]")[1:10],], type="html") ``` <table> <thead> <tr> <th style="text-align:left;"> ensembl75_id </th> <th style="text-align:left;"> gname </th> </tr> </thead> <tbody> <tr> <td style="text-align:left;"> ENSG00000005955 </td> <td style="text-align:left;"> GGNBP2 </td> </tr> <tr> <td style="text-align:left;"> ENSG00000006074 </td> <td style="text-align:left;"> CCL18 </td> </tr> <tr> <td style="text-align:left;"> ENSG00000006075 </td> <td style="text-align:left;"> CCL3 </td> </tr> <tr> <td style="text-align:left;"> ENSG00000006114 </td> <td style="text-align:left;"> SYNRG </td> </tr> <tr> <td style="text-align:left;"> ENSG00000017621 </td> <td style="text-align:left;"> MAGIX </td> </tr> <tr> <td style="text-align:left;"> ENSG00000034063 </td> <td style="text-align:left;"> UHRF1 </td> </tr> <tr> <td style="text-align:left;"> ENSG00000049319 </td> <td style="text-align:left;"> SRD5A2 </td> </tr> <tr> <td style="text-align:left;"> ENSG00000056661 </td> <td style="text-align:left;"> PCGF2 </td> </tr> <tr> <td style="text-align:left;"> ENSG00000068793 </td> <td style="text-align:left;"> CYFIP1 </td> </tr> <tr> <td style="text-align:left;"> ENSG00000069712 </td> <td style="text-align:left;"> KIAA1107 </td> </tr> </tbody> </table> --- ## Assignment #1 * <font size=5> Due February 4th, 2020 @ 20:00 </font> ## What to hand in? * **html rendered RNotebook** - you should be able to submit this through quercus * Make sure the notebook and all associated code is checked into your github repo as I will be pulling all the repos at the deadline and using them to compile your code. - Your checked in code must replicate the handed in notebook. * **Do not check the data file into your repo!** - your code should download the data from GEO and generate a new, cleaned data file. * Document your work and your code directly in the notebook. * **Read the paper associated with your data!** * You are allowed to use helper functions or methods but make sure when you source those files the paths to them are relative and that they are checked into your repo as well.